Capillary electrophoresis DNA sequencers are used to determine the sequence of bases in DNA, the blueprint of life, as well as the length of DNA fragments. These sequencers are in widespread use around the world in medicine, healthcare, and criminal investigation including DNA profiling. To supply the market for compact models that is expected to grow strongly in the future, Hitachi High-Tech Corporation has developed the DS3000 sequencer that is easy to use and suitable for the small-lot-size testing of diverse samples. The new sequencer has been launched on the Japanese and Asian markets. This article describes the DS3000 sequencer and its features, which include the adoption of a direct polymer injection mechanism and reagent cartridges for rapid analysis when used for small-lot-size testing of diverse samples, space-efficiency achieved through use of a panel computer and laser diode, and a remote monitoring system that significantly improves usability.





The foundations of today’s genomic medicine were laid by the Human Genome Project of the early 2000s. The capillary electrophoresis (CE) deoxyribonucleic acid (DNA) sequencers (hereinafter “CE sequencer”) developed by Hitachi made a significant contribution to this project by increasing the efficiency of the gold-standard Sanger sequencing method. The market for DNA sequencers grew considerably following the emergence from around 2005 of next-generation sequencers that worked by the massively parallel analysis of short DNA fragments, and a number of benchmark DNA databases (reference sequences) have since been established. Meanwhile, markets for specific applications in fields such as medicine and criminal investigation that involve the analysis of individual DNA differences have been expanding since about 2015.
CE sequencers are also used for forensic DNA profiling(1), where they take advantage of their ability to perform simple and low-cost analyses of particular sections of DNA. They also have an important role to play in healthcare such as in molecular diagnostics, where one application is the detection of microsatellite instabilities (MSIs)(2), genetic abnormalities that can help identify which therapies to use on solid carcinomas such as colon cancer.
Strong growth is anticipated in the market for compact desktop sequencers with four or fewer capillaries. Working in collaboration with Promega Corporation, a company with expertise in the development and production of reagents, Hitachi High-Tech Corporation has developed a compact CE sequencer that is easy to use and suitable for small-lot-size testing of diverse samples. The companies have launched the new sequencer as the Hitachi-branded DS3000 in the Japanese and Asian markets, and under the Promega brand in the rest of the world. Promega operates a global sales and service network(3), (4).
The demand for large high-throughput CE sequencers decreased significantly following completion of the Human Genome Project in 2003, with the emergence of next-generation sequencers also a factor. However, with CE sequencers finding widespread use in applications for medicine, healthcare, and criminal investigation such as DNA profiling at universities and other academic institutions or at government-run research institutes, there has been steady market demand for compact desktop sequencers intended for laboratory use in particular.
The compact sequencer market is characterized by the diversity of samples to be analyzed and the wide variety of applications, which include DNA sequencing and fragment analysis (measurement of the length or quantity of specific sections of DNA) (see Figure 1). This creates a need for instruments suitable for small-lot-size testing of diverse samples, with a user interface appropriate for non-experts and small enough to be installed in the confined space of a laboratory. Moreover, rather than contracting sequencing work out to a specialist provider or using a shared in-house laboratory, there is also demand from customers who want to have their own machine in their own laboratory, perhaps because they require rapid results or have stringent requirements for results handling. While this led to the Applied Biosystems 310 Genetic Analyzer (a single-capillary sequencer) becoming widely used, the model was discontinued in 2011 leaving a need for an alternative compact sequencer.
To meet the needs of users in this market, Hitachi High-Tech launched a newly developed compact model with exceptional ease-of-use and the ability to switch between different reagents (polymers) quickly and easily for different analytical purposes.
Figure 1 — Use of Different Reagents (Polymers) for Different Types of Analysis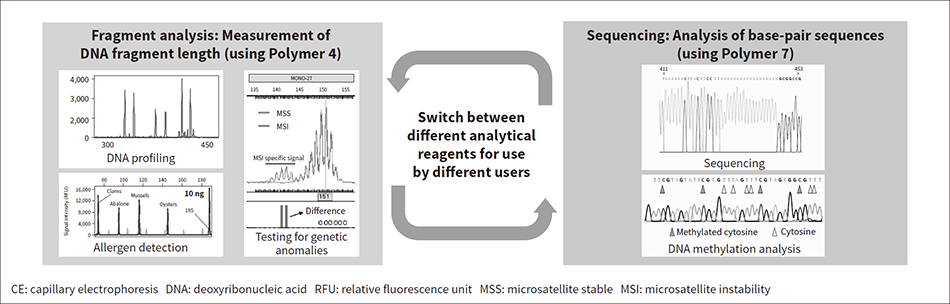 CE sequencers use different polymers to perform different types of analysis. A sequencer may have a number of users and have to alternate between performing fragment analysis (DNA profiling or testing for genetic anomalies) and DNA sequencing.
CE sequencers use different polymers to perform different types of analysis. A sequencer may have a number of users and have to alternate between performing fragment analysis (DNA profiling or testing for genetic anomalies) and DNA sequencing.
Figure 2 — DS3000 Sequencer and Consumables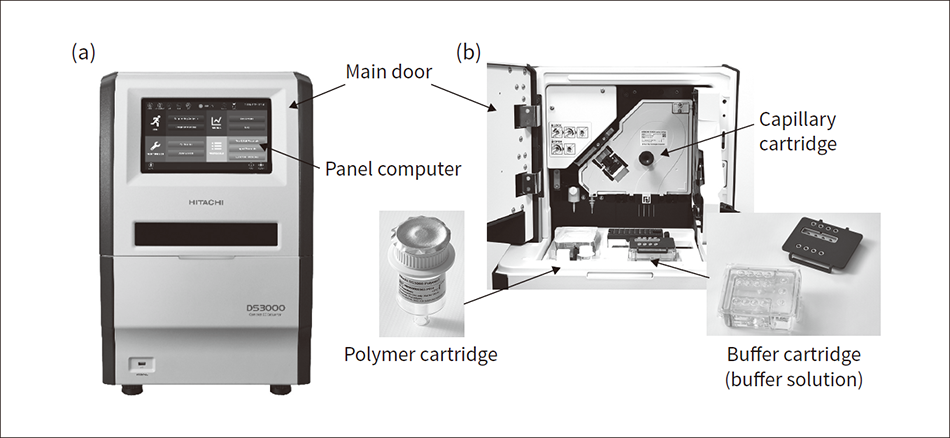 The figure shows the sequencer unit (a) and an interior view with the main door open that also shows the consumables and where they are inserted (b). The panel computer that controls the sequencer is built into the front-side door. The consumable reagents are supplied in cartridges (disposable containers) and fitting the capillary array is also a simple task.
The figure shows the sequencer unit (a) and an interior view with the main door open that also shows the consumables and where they are inserted (b). The panel computer that controls the sequencer is built into the front-side door. The consumable reagents are supplied in cartridges (disposable containers) and fitting the capillary array is also a simple task.
For the DS3000, Hitachi High-Tech developed a direct polymer injection mechanism and new polymer cartridges to meet the needs of users wanting to conduct a variety of different measurements (see Figure 2). The new sequencer also featured an integrated panel computer for user operation and primary analysis, ensuring small size and keeping installation space requirements to a minimum. A remote monitoring function was also developed that works by connecting the sequencer to the user’s network to enable the collection of measurement data and the monitoring of instrument operation from a PC. This section describes these product features.
Figure 3 — Comparison of Polymer Transportation Mechanisms and Transportation Paths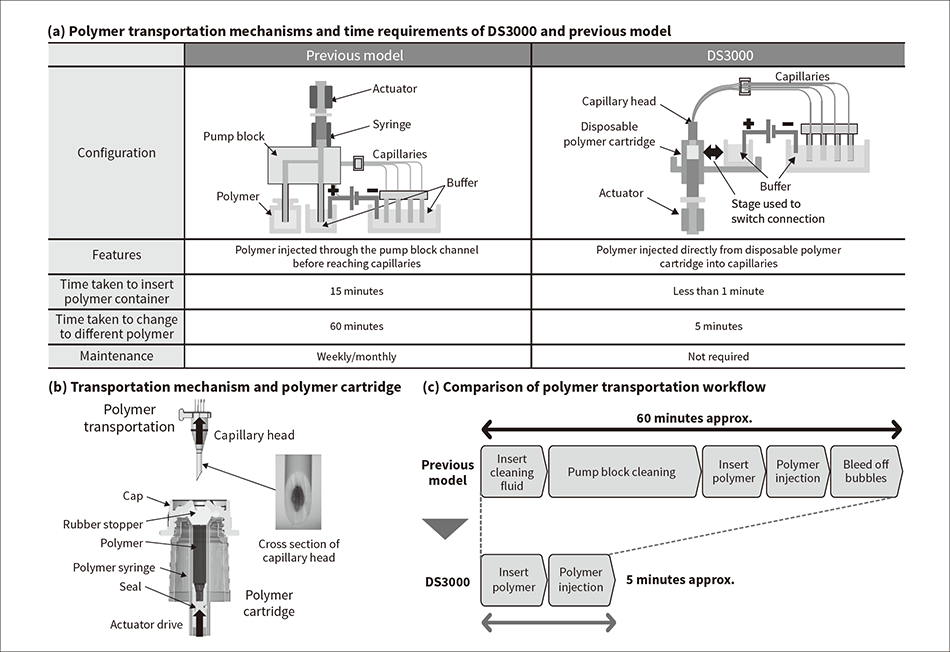 The polymer transportation system for the DS3000 is made up of a polymer cartridge and an actuator. The polymer cartridge is a consumable item that also works as a syringe. The actuator pushes up on the bottom of the polymer cartridge to deliver polymer to all four capillaries at once. The new mechanism significantly reduces the time and effort required to change to a different polymer.
The polymer transportation system for the DS3000 is made up of a polymer cartridge and an actuator. The polymer cartridge is a consumable item that also works as a syringe. The actuator pushes up on the bottom of the polymer cartridge to deliver polymer to all four capillaries at once. The new mechanism significantly reduces the time and effort required to change to a different polymer.
The main intended users of the sequencer are teams of researchers working in small laboratories engaged in things like clinical research or DNA profiling who share use of the instrument. What these users need, then, is an easy way to inject polymer into the capillaries as required for their particular purposes, whether it be DNA sequencing, fragment analysis, or something else.
Unfortunately, past models have required about 60 minutes to switch between polymers. The reason why this took so long was due to the need to replace the polymer in the flow channels and to bleed the pump block channels to eliminate bubbles.
To overcome this problem, Hitachi High-Tech developed a new polymer injection system that eliminates the pump block, instead using an actuator that connects the disposable polymer cartridge directly to the capillary (see Figure 3). The new system does not have to be bled to eliminate bubbles and makes it easy to switch to a different polymer as needed by the application. Also, the syringe function that used to be part of the sequencer unit is now built into the polymer cartridge. As this was an area prone to being clogged by polymer crystallization, incorporating it into a consumable part eliminates the need for customer maintenance to clean out the polymer injection mechanism and associated parts.
Figure 4 — Network Connection and Remote Monitoring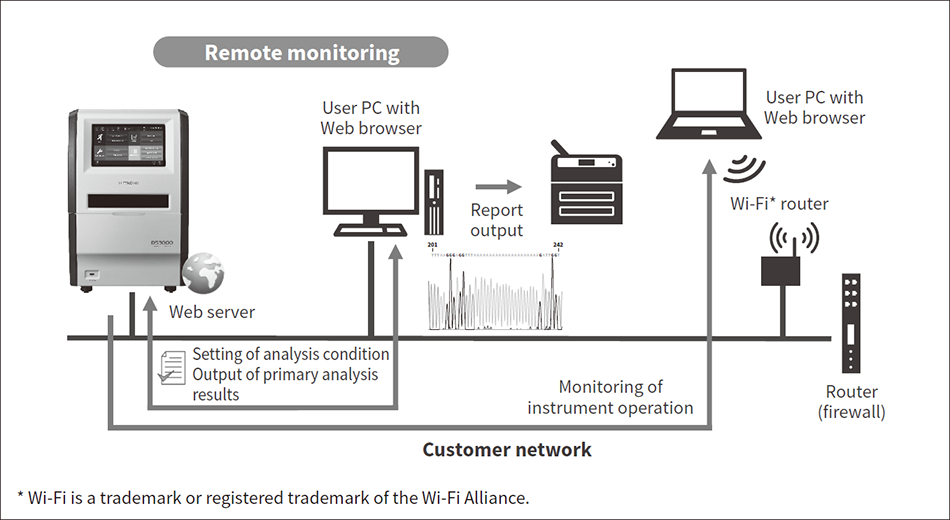 Accessibility is improved by functions for remote monitoring of the instrument during analyses, including real-time display of the electrophoretogram, and for downloading measurement results and reports from a browser.
Accessibility is improved by functions for remote monitoring of the instrument during analyses, including real-time display of the electrophoretogram, and for downloading measurement results and reports from a browser.
Figure 5 — Instrument Miniaturization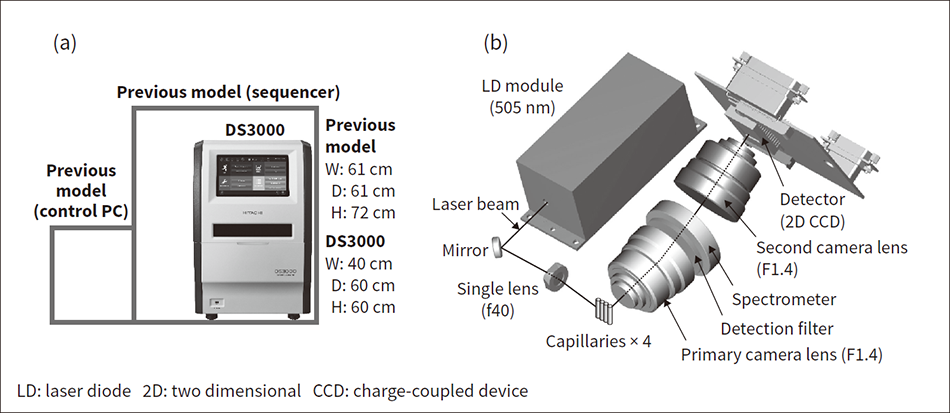 The image on the left (a) compares the size of the DS3000 to the previous model and that on the right (b) shows the miniaturized optics. The new optics and integration of a panel computer succeeded in reducing the size of the instrument and making more efficient use of space.
The image on the left (a) compares the size of the DS3000 to the previous model and that on the right (b) shows the miniaturized optics. The new optics and integration of a panel computer succeeded in reducing the size of the instrument and making more efficient use of space.
The new sequencer takes up only about 60% as much space as the previous model. This was achieved by integrating the panel computer used for instrument operation and sequencing of the measurement results and by miniaturizing the optics through the use of a laser diode light source (see Figure 5).
A new 505-nm semiconductor laser diode (LD) was adopted to reduce the size of the optics system. In contrast, the previous model used a diode-pumped solid-state (DPSS) light source. The DPSS was made up of the controller and a laser head unit to generate a continuous-wave from a laser crystal pumped by a laser diode as the excitation source. As a mechanical shutter was also used to block excitation light during charge-coupled device (CCD) data transmission, considerable space was required. On the new system, in contrast, the system size has been reduced by the laser beam pulsed from the LD, the single module combining laser head and controller, and the shutterless design.
This section presents example uses of the sequencer to analyze a variety of different samples. As each of these analyses requires different polymers and sequencer settings, they provide a practical demonstration of the DS3000’s features and its ability to handle different sample types (see Table 1). Use of the sequencer for DNA profiling(5) and in medical research have also been reported elsewhere.
Table 1 — Instrument Settings for Different Types of Analysis The DS3000 makes it easy to change the analytical reagents (polymers) and settings (assay and number of fluorescent dyes) shown in the table.
The DS3000 makes it easy to change the analytical reagents (polymers) and settings (assay and number of fluorescent dyes) shown in the table.
Genomes contain regions called microsatellites where a distinctive DNA sequence of between one to six base pairs is repeated several dozen times. Anomalies in these sections of DNA are called microsatellite instabilities (MSIs) and are known to be biomarkers predictive of defective mismatch repair proteins. As MSI analysis involves the detection and comparison of dispersed waveform peaks at the level of single base pairs, this is an application that demands a high level of sizing accuracy and replicability. This section describes an MSI analysis conducted on commercially available paraffin-embedded samples.
The samples in this case were a reference sample with a known MSI (MSI, HD830)*1 and a wild-type (non-mutated) sample [microsatellite stable (MSS) HD831]*1. The microsatellite region was amplified using Promega MSI Analysis System Ver. 1.2 and electrophoresis was performed using the resulting amplification product on Polymer 7.
Figure 6 (a) shows the results of analyzing the waveform data. This shows that the analysis has successfully detected all of the distinctive MSI peaks(6), (7).
Low-frequency mutations are mutations present in a sample derived from multiple cells that occur in only a subset of those cells. Such mutations are recognized for their utility in cancer research as they can be a cause of the initiation and growth of tumors. As the signal from these low-frequency mutations is buried amid that of the wild-type DNA that makes up the bulk of the sample, their detection requires that sequencing have a high level of sensitivity and accuracy. This example describes the use of a DS3000 to detect low-frequency mutations in the KRAS proto-oncogene.
A genetic analysis reference sample was used for the demonstration*1. The sample contained DNA with a known number of mutations in specific sections, and these sections and frequencies were open to the public. The test sample containing low-frequency mutations was created by diluting this reference sample with wild-type DNA. Sequencing was then performed targeting the KRAS gene. The resulting waveform data was analyzed using Mutation Surveyor v5.1.2 (from SoftGenetics, LLC). Figure 6 (b) shows an example result in which a low-frequency mutation present in 10% of cells was successfully detected*2.
Between 1 to 3% of adults and 4 to 6% of children suffer from food allergies, with severe reactions in certain cases that can be life-threatening. This makes allergen detection and labeling an important food safety issue. This example describes a highly sensitive detection technique that uses a combination of multiplex polymerase chain reaction (PCR) and capillary electrophoresis to target the gene for tropomyosin, a known allergen widely present in shellfish(8).
First, DNA was extracted from commercially purchased shellfish (clams, abalone, mussels, and oysters) and mixed in equal proportions. Next multiplex PCR was used to amplify the tropomyosin gene and 18S rDNA, the latter being used as a positive control. Electrophoresis was performed using Polymer 4 on the resulting product. The results are presented in Figure 6 (c), showing a waveform peak that corresponds to a tiny 0.01 ng of shellfish DNA(9).
Figure 6 — Analyses Conducted Using DS3000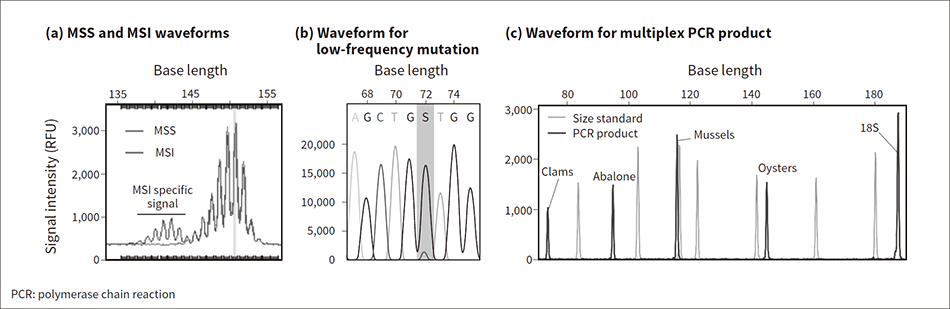 The graph in (a) overlays the MSS and MSI waveforms. The peak waveform was plotted using GeneMarker v3.0.1 (from SoftGenetics, LLC). The graph in (b) shows the waveform for a KRAS mutation present at low frequency. Mixed base (S) indicates a guanine (G: peak in black) overlapping with cytosine (C: peak in grey) of which the frequency is 10%. The sensitivity of low-frequency mutation detection depends on the condition of the sample and the sequence contexts. The result presented here is an example obtained under restricted conditions. The graph in (c) shows the waveform for the multiplex PCR product obtained from 0.01 ng of shellfish genome DNA (using SoftGenetics GeneMarker HID v2.9.3).
The graph in (a) overlays the MSS and MSI waveforms. The peak waveform was plotted using GeneMarker v3.0.1 (from SoftGenetics, LLC). The graph in (b) shows the waveform for a KRAS mutation present at low frequency. Mixed base (S) indicates a guanine (G: peak in black) overlapping with cytosine (C: peak in grey) of which the frequency is 10%. The sensitivity of low-frequency mutation detection depends on the condition of the sample and the sequence contexts. The result presented here is an example obtained under restricted conditions. The graph in (c) shows the waveform for the multiplex PCR product obtained from 0.01 ng of shellfish genome DNA (using SoftGenetics GeneMarker HID v2.9.3).
This article has described Hitachi High-Tech’s DS3000 compact CE sequencer, which is suitable for small-lot-size testing of diverse samples.
Along with seeking to identify customer issues and to foster and expand the market for CE sequencer applications that take advantage of advanced technological developments, Hitachi also intends to contribute to the creation of a safe and healthy society through the operation of its digital solutions business that integrates data information from such diverse sources as genomics, medicine, health, and the environment.