Enabling rapid search of genomic databases of totaling 45.7 million records.
HiRDB, a parallel RDB to support development of applied genome science.
With the renewal of their supercomputer system in January 2003, the Human Genome Center at the Institute of Medical Science, The University of Tokyo, updated their publicly available Internet service, which allows any user to search genomic databases. The center chose HiRDB, a scalable database from Hitachi, to achieve rapid searching of the genome data, which consists of 82.6 billion characters and 45.7 million records (as of April, 2005). HiRDB offers high-speed response, due to its scalable parallel processing and flexible data storage. Highly extensible HiRDB efficiently stores the increasing amount of genetic information to support the continuing development of applied genome science.
"With the help of scientists across the world, the Human Genome Project announced in 2003 the completion of its goal: to determine the entire human DNA sequence," remarked Dr. Kenta Nakai, professor at the Laboratory of Functional Analysis in silico, for the Human Genome Center at the Institute of Medical Science, The University of Tokyo. "Now, we're entering an era in which the information we've gained will be put to good use in medicine."
A genome is a complete set of genetic information. Genomes consist of DNA, which encodes genetic information as a sequence of four types of bases (or letters). The number of letters determined worldwide reached 82.6 billion on April, 2005. However, the task of analyzing the various roles of these sequences, as well as functions of the sequences when combined, still lies ahead.
"By taking advantage of this new information, we can achieve 'personal medicine', which knows what medicines are most effective for any given individual, " Dr. Nakai adds. "We'll have a better academic understanding of the design principles of genetic information, which will most likely lead to increased efforts to zero in on the essence of life."
The Human Genome Center at the Institute of Medical Science, The University of Tokyo, was founded as a focal point of Japan's genome analysis, when plans for the Human Genome Project were launched in 1991. It is also a center of Japan's international contribution and international competition for genome research.
A high-speed supercomputer system plays a vital role in helping researchers across a wide variety of fields, including biologists, geneticists, and researchers in experimental medicine, aiming to achieve tailor-made medicine and personal medicine, to find the sequence information they seek from the vast amount of sequence data. In January 2003, the Human Genome Center reconfigured their supercomputer system to a system consisting of more than a dozen UNIX machines. Since the amount of data continues to grow daily at an extremely rapid pace, SANRISE, used to store data for the supercomputer system, provides roughly 150 TB of capacity.
"Perhaps it would be efficient to have one conventional supercomputer when performing a vast amount of specific mathematical calculations," explains Dr. Nakai regarding the goals of the reconfiguration, "but the human genome consists of character information regulated by an unknown grammar, not mathematical information. Also, our system needs to provide a variety of services, such as database searching, genome analysis, sequence analysis, simulation, and joint research. As such, load balancing is an important part of the reconfiguration.
"As the amount of information we obtain increases, so does the number of types of analysis, as well as the diversity of what we want to do," adds Mr. Toshiaki Katayama, Research Associate of the Human Genome Center at the Institute of Medical Science, The University of Tokyo.

Human Genome Center, Institute of Medical Science, The University of Tokyo
Professor, Laboratory of Functional Analysis in silico
Mr. Kenta Nakai

Human Genome Center, Institute of Medical Science, The University of Tokyo
Assistant, Genome Database
Mr. Toshiaki Katayama
With the reconfiguration of their supercomputer system, the center started a service called the "HiGet System". This is a public, Internet-based search service that provides genomic databases access to anyone.
HiGet System search screenshot
See larger picture in a new window
Usually, accession numbers are used to reference genetic sequence data in academic papers. If researchers reading such a paper want to know more about the sequence of the gene corresponding to the accession number, they can go online immediately to access the HiGet system, and use the accession number to search for the sequence. Alternatively, if researchers want to know more about what genes are found in a species that they are researching, the researcher can use a conditional search by keyword to list the corresponding genes. When more can be done using genetic information, the user base will grow as users will want to accomplish more, and the more important the HiGet system will become. "An Internet-based search service for genomic databases is not necessarily something new, but since this service allows more flexibility than conventional services in using search conditions to narrow down results, users can find the information they want in less time," explains Mr. Katayama.
For the HiGet system database, the center uses HiRDB, a scalable database from Hitachi. To enable high-speed searches for the HiGet system, Hitachi drew on its complete experience in supercomputer systems, performing system-wide optimization by unifying the hardware, database and application components, such as the CPU and I/O interface. Hitachi also paid close attention to the system design, using a large index buffer to improve search speed. In addition, the Platform Systems Research Department of Hitachi's Central Research Laboratory, which performs research in database systems, teamed up with Hitachi's Life Science Group, using the group's academic knowledge of genomes and DNA to design the hierarchy of search tags.
Hitachi system engineers also played a role, using their detailed understanding of database structures as well as the fast support and geographical advantage of a domestic manufacturer to maintain a high level of system reliability and availability. As such integration is Hitachi's forte, it was successful in setting up a key database system to support genome research.
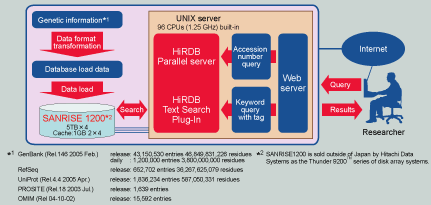
Because the HiGet system provides an important database service around the clock to users across the globe, it consists of two systems: data on one side can be used for additions and reconfiguration, while the data on the other side can be used for the search service. As such, once data is added or reconfigured on the former database, the search target database can then be switched to the former database. The platform brings out maximum performance for the HiRDB parallel RDBMS, by using a high-end UNIX parallel server with 96 1.25GHz processors built in. Even though HiRDB consists of 82.6 billion characters and an impressive 45.7 million records of genetic information, it returns quick responses to Internet queries.
HiRDB's parallel processing utilizes a "shared nothing architecture". With this method, since there are almost no shared resources, the parallel processing power really excels as the number of CPUs increases. With genomic databases, data is stored across 32 areas, and HiRDB's parallel search is used to achieve unprecedented search speeds.
With HiRDB, data storage can be designed flexibly. Partitioning/parallel search is such an example, since it allows an increase in the number of database records, without affecting the speed of searches by accession number.
And with HiRDB's full text search engine, the HiRDB Text Search Plug-In, full text search indexes can be stored as-is in a database field. This means that even processing for complicated keyword search conditions that include AND, OR, and NOT operands can be performed via closed logical calculations within the database, enabling fast responses.
Users are satisfied that responses are faster," explains a confident Dr. Nakai. "Performance for both accession number searches and condition searches is now as good as, if not better than, that for famous database search services in the U.S."
The performance that only a parallel database could offer excels even under daily data load.
On the public sites in the U.S. and the U.K., around 1,200,000 records of new genetic information are provided daily, or around 43,150,530 records adjusted over twomonth increments. The HiGet system currently consists of 82.6 billion characters and 45.7 million records of genetic information (as of April, 2005), and is updated daily. The data import that takes place daily and every two months is performed quickly, using HiRDB's parallel loading.
Now, as usage grows for the HiGet system, demands for the service will diversify.
"In addition to genomic databases, we're planning more complex searching, such as lateral searching in a protein database," says Dr. Nakai. "Then we'll add new types of search and analysis, but they too will surely be based on the HiGet system and HiRDB." With its ability to deliver fast search performance even after system extension, HiRDB is a perfect fit for supporting the development of Japan's genome research.
Human Genome Center HiGet system overview
See larger picture in a new window
User Profile