Capillary array DNA sequencer
The advent of molecular biology in the 1980s and progress on the Human Genome Project in the 1990s, gave rise to the need for a DNA sequencer capable of decoding base pairs of large quantities of DNA with a high degree of precision and speed. The first DNA sequencers, known as"slab gel" sequencers, consisted of a gel substance sandwiched between two flat panels of glass through which DNA fragments were migrated via electrophoresis. Capillary array DNA sequencers, where DNA fragments undergo electrophoretic migration through narrow, gel-filled glass capillaries, were also developed. Both these early types of DNA sequencers were marred by poor fluorescence detection and inefficiency, requiring a laser to be directly beamed sequentially through each capillary in the array.
Through a number of technological innovations leading to the "Sheath-flow" technique, Dr. Kambara achieved across-the-board improvements in the functionality of the device. These innovations were particularly vital in preventing the scattering of laser light irradiated across the surface of the array, the most vexing issue surrounding parallel analysis in capillary arrays at the time. With this technique, Dr. Kambara succeeded in simultaneously detecting fluorescence in an array of 96 capillaries, while raising the sensitivity of fluorescence detection by more than an order of magnitude. Analytical precision was then vastly increased by fully automating control of the entire process.
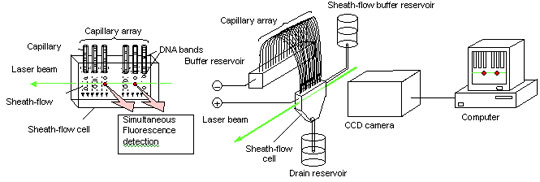
Capillary Array DNA Sequencer using multiple Sheath-flow method
Dr. Kambara further discovered that the process could be sped-up by removing much of the heat arising from the electric field used in DNA analysis by adjusting the diameter of, and the distance between individual capillaries, and by air cooling the capillary walls. The end result was a high-throughput capillary array DNA sequencer with a functional capacity nearly 20 to 30 times that of previous devices.
This revolutionary device was soon drafted for use in the Human Genome Project, where it led to groundbreaking results. To make the device viable for use in ordinary research laboratories, Dr. Kambara also developed a "Multiple-Focusing Irradiation" technique that further simplified construction of the sequencer. This new design not only improved device control and maintenance, but made low-cost capillary array DNA sequencers well suited to general purpose tasks a reality.
Development of this latest DNA sequencer has resulted in a fully automated method for decoding DNA base pairs with a level of fluorescence detection sensitivity and throughput capacity more than an order of magnitude higher than previous devices.
Aside from its decisive contribution to the Human Genome Project, this new sequencer is encouraging the advancement of new research in biology and biochemistry, as well as the development of scientific technology in pharmacology, medicine and other research fields utilizing applied genomic data.
Background
The advancement of recombinant DNA technologies and techniques for decoding DNA base pairs in the field of molecular biology in the 1980s, gave a dramatic boost to research into analyzing and understanding life on the molecular level. The appearance of automated equipment for tagging DNA with fluorescent markers was a major factor in this progress, replacing radioactive isotope markers. As the international Human Genome Project got underway in the 1990s, demand grew for high-throughput DNA analysis equipment capable of outperforming conventional technology by at least a single order of magnitude. Attention focused on developing a fully automated fluorescent DNA sequencer as a solution for meeting this demand. Research and development in DNA analysis and other areas continued to move forward, with successful research finding useful applications in medicine and other fields. The widening range of applications, in turn, began to fuel rapidly growing demand for low-cost, easy-to-use DNA sequencers.
At the time, both "slab gel" DNA sequencers and early capillary array DNA sequencers were already available. Early versions of the latter, however, were marred by poor fluorescence detection and inefficiency, requiring a laser to be directly beamed sequentially through each capillary in the array. Thus it was hoped that a fully automated, easy-to-operate DNA sequencer, with a high degree of precision and high throughput, would one day soon be developed.
Development
Determined to satisfy this need, Dr. Kambara focused his attention on the capillary array DNA sequencer. Involved in research and development in the genetics field since capillary array DNA sequencers were first proposed, Dr. Kambara was convinced that performance enhancements such as high-speed electrophoresis, parallel DNA sequencing and more sensitive fluorescent detection held the key to realizing high-throughput DNA base reading, based on his research experience in genetic analysis. Dr. Kambara developed a high-performance DNA sequencer with a fluorescence detection system using the fundamental technologies explained below.
1) Side-Entry Laser Irradiation Technology
DNA sequencers were put to use for the first time in 1986, in the U.S. The original slab gel DNA sequencers used a scanning laser-beam irradiation method where the laser beam was aimed at different positions on the slab gel sequentially. Repeated scans of irradiation positions allow the photo-detector to measure fluorescence emissions from multiple electrophoresis migration paths in order, and determine the base sequence of multiple DNA samples, however, time-sharing fluorescence detection using laser scans in this manner also suffered several drawbacks.
To address these and other issues, Dr. Kambara developed a slab gel DNA sequencer with "side-entry laser irradiation" technology (Fig. 1). The laser enters the slab gel from the side and traverses the gel medium between the two panes of glass. This arrangement allows the laser beam to simultaneously intersect all electrophoresis migration paths, thereby increasing irradiation efficiency. A multi-pixel array CCD camera simultaneously detects fluorescence emissions from each migration path. This technology allows fluorescence emissions from numerous DNA samples to be detected at the same time, without the need for fluorescence measurements. This technology succeeded in increasing detection sensitivity by more than ten times that of DNA sequencers using conventional laser scans. Devices using this technology made their commercial debut as Japan's first slab gel DNA sequencers in 1989.
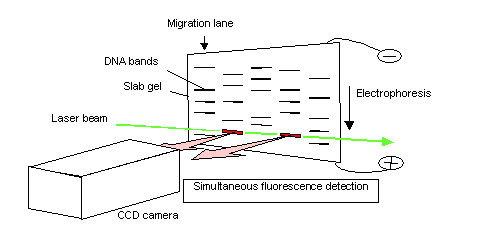
Fig. 1. Slab gel DNA sequencer using side-entry laser-beam irradiation
2) Sheath-Flow Technology
While the Human Genome Project was still on the drawing board, Dr. Kambara was one of the first to realize that the success of the project would depend upon the development of a DNA sequencer that could rapidly sequence bases with a high throughput. In 1989, this belief prompted Dr. Kambara to begin developing a DNA sequencer equipped with next-generation technologies that would realize high speed and throughput. Of the many alternative technologies, Dr. Kambara demonstrated that capillary array DNA sequencers represented the most promising and effective technology for high-speed DNA sequencing.
Gel-filled capillaries, which replace the slab gel and act as the electrophoresis migration path for DNA fragments, dissipate heat much more effectively than conventional slab gels. This means that capillaries can withstand the higher applied voltages that are required to accelerate electrophoresis. Another promising advantage of gel-filled capillaries over slab gels is the high throughput made possible by parallel sequencing of large capillary arrays. Each capillary represents one electrophoresis migration path, and can be used for base reading one DNA sample. However, a difficulty associated with this approach is that laser beams are scattered and refracted by capillaries.
The most pressing research topic, therefore, was to develop a method of laser irradiation which would allow the laser beam to penetrate numerous capillaries. Two university research groups in the U.S. developed their own laser irradiation methods in response to this problem. A research group at the University of California developed a laser scanning method (Fig. 2). With this method, a bundle of capillaries are placed on a flat surface and laser beams are directed at single capillaries in sequence to induce fluorescence emission for detection. As shown in Figure 2, the capillary array is repeatedly scanned by the laser beam, one capillary at a time. Each capillary is irradiated with the laser and fluorescence is detected. In this manner, multiple DNA samples are sequenced.
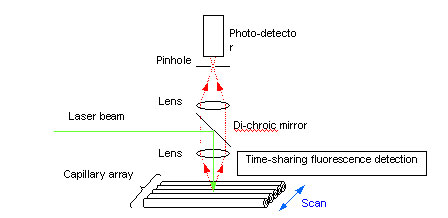
Fig. 2. Capillary array DNA sequencer using scanning laser-beam irradiation
One of the drawbacks of time-sharing laser-induced fluorescence detection, however, is that an increase in the number of capillaries shortens the time available to detect fluorescence emissions from each capillary, and thus from each DNA sample. This in turn lowers the sensitivity of fluorescence detection. Another drawback is that scanning speeds may be unable to keep pace with faster electrophoresis migration speeds. Further, the moving components of the scanning mechanism resulted in a very large apparatus and lead to maintenance issues due to the greater likelihood of malfunction.
The second research group that sought to improve laser irradiation efficiency, based at Iowa State University, developed a laser irradiation method that relied on beam-width expansion (Fig. 3).
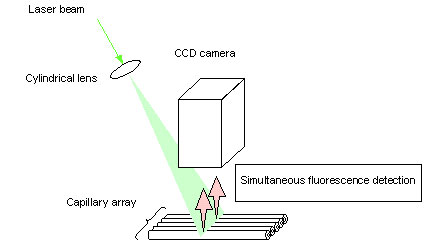
Fig. 3. Capillary array DNA sequencer using expanding laser-beam irradiation
Many capillaries are arranged on a flat surface and the laser beam is expanded in the forward direction of the capillary array plane, irradiating all capillaries at once. A CCD camera is used to simultaneously detect fluorescence emission from all capillaries. Under this method, the intensity of the laser beam decreases as the beam width is expanded. This means that the total intensity of irradiation per capillary will decrease in proportion to an increase in the total number of capillaries being sequenced in parallel. The result is that the fluorescence emission per DNA sample will decrease, lowering the sensitivity of fluorescence detection.
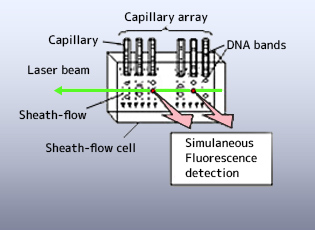
In response to these issues, Dr. Kambara formulated an original technique called the "sheath-flow" method (Fig. 4) on the premise of his research experience in slab gel; that the most efficient irradiation method is to irradiate all capillaries simultaneously by introducing the laser from the side of the capillary array - the direction perpendicular to the capillary array plane. In this geometry, however, the laser beam is scattered and refracted by the capillary tubes. To avoid this problem, the ends of the capillaries, which act as migration paths, were immersed in a sheath fluid (buffer solution) inside a sheath-flow cell. The DNA fragments are irradiated from the side by the laser and the signal emissions are detected as they are eluted from each capillary. The sheath fluid is introduced into the sheath-flow cell from a supply tank, and is discharged from the sheath-flow cell into a disposal tank.
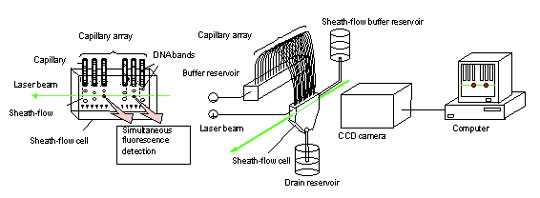
4. Capillary Array DNA Sequencer using Multiple Sheath-flow
Sheath-flow technology removes obstacles from the irradiated area, giving several key advantages: simultaneous laser irradiation of all migration paths, sharp reductions in background light, and simultaneous detection of fluorescence from all migration paths by a multi-pixel array CCD camera. The technology does not rely on the time-sharing laser irradiation and signal detection required by laser scans, or on the spatially distributed laser irradiation characteristic of the expanded laser beam method. Consequently, the technology effectively yields the same degree of fluorescence detection sensitivity as would be obtained by illuminating the laser on a single capillary continuously. In fact, the fluorescence detection sensitivity of sheath-flow technology is more than ten times greater than methods that use laser scans or expanded laser beams.
A paper on sheath-flow technology was published in the scientific journal 'Nature' in 1993. The sheath-flow technology was licensed to the U.S. in 1998, and used to commercialize a high-throughput capillary array DNA sequencer. This apparatus sequences 96 capillaries in parallel, with a throughput 20 to 30 times greater than conventional slab gel DNA sequencers. Further, capillary gel refilling and DNA sample introduction procedures were fully automated to yield both labor and cost savings.
3) Multiple-Focusing Irradiation Technology
Although the sheath-flow method is ideal for the high-throughput capillary array DNA sequencer, as sequencers became larger and more complex, maintenance of the sheath-flow apparatus became more difficult and costly. While capillary array DNA sequencers have made a significant contribution to the progress of genome projects, and the data is now being used in a variety of contexts to analyze base sequences and perform comparative genetic analysis for species, it is also being widely used by industry in fields such as drug discovery, DNA diagnostics, and the development of micro-organisms for industrial use. As such, the need for quality DNA sequencers is no longer exclusive to specialized genome analysis centers, and a need for easily operated low-maintenance instruments will soon be required. This issue compelled Dr. Kambara to begin developing an inexpensive and simplified capillary array DNA sequencer in 1994, resulting in an enhanced method of laser irradiation called the " multiple focusing irradiation " method (Fig. 5). Specifically, Dr. Kambara determined the conditions under which capillaries would behave as convex lenses. These conditions required adjusting the ratio of the inner and outer diameters of capillaries, refractive indexes and other parameters, and were determined by extensive analysis of the reflection and refraction properties of laser beams incident upon gel-filled capillaries. Under these conditions, a laser beam entering a capillary array from the side can be focused repeatedly by capillaries. The result is that all capillaries can be irradiated efficiently and fluorescence emission from every capillary can be simultaneously detected by a CCD camera. Furthermore, highly simplified DNA sequencers can be realized using side-entry multiple-focusing irradiation, because this method does not require a complex sheath-flow mechanism
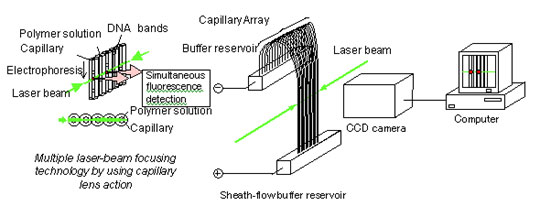
Fig. 5. Capillary array DNA sequencer using multiple laser-beam focusing
A paper on this technology was published in the scientific journal 'Analytical Chemistry' in 1996. The technology was used to commercialize a general-purpose capillary array DNA sequencer in 2000.
By automating all processes and simplifying the structure of the optical measurement apparatus, the instrument can be more flexibly configured for use. The new design also succeeded in lowering the price of the instrument, made it easier to operate and reduced maintenance requirements. The instrument also allows users to flexibly adjust capillary length, electrophoresis voltage and temperature settings.